Page 83 - 2025S
P. 83
76 UEC Int’l Mini-Conference No.54
Swin-UNet++: A Multi-Scale Transformer-CNN Hybrid for Brain
Tumor Segmentation in MRI Scans
Rinvi Jaman Riti*, Norihiro Koizumi, Yu Nishiyama, Peiji Chen
UEC Exchange Study Program (JUSST Program)*
Department of Mechanical and Intelligent Systems Engineering
The University of Electro-Communications, Tokyo, Japan
Introduction Implementation Details
Brain tumors are the most critical conditions Lists software tools, training setup, and computational
requiring precise diagnosis and treatment planning configuration:
[1]. Manual segmentation of brain tumors from MRI
Framework PyTorch
scans is time-consuming and subject to inter-
observer variability. Deep learning methods, Loss Dice + Cross-Entropy
especially convolutional neural networks (CNNs), Optimizer Adam
have shown promising performance in automating Learning Rate 0.0001 with scheduler
segmentation [2]. However, CNNs often struggle Epochs 100–150
with capturing global context. Swin-UNet++ Augmentations Rotation, flipping, normalization
combines the strengths of CNNs and Transformer
architectures to deliver a multi-scale, attention-
enhanced segmentation pipeline. Evaluation Metrics Comparison
Evaluation metrics used to validate segmentation:
Dataset
Table 1: Evaluation metrics comparison
Brain tumors public BRATS 2021 dataset [3] collected Dice
Model IoU Notes
from Kaggle. For 3 categories: tumor, edema, necrotic Score
core. Total number of images: 1251
UNet 0.82 0.75 Good local features
UNet++ 0.85 0.78 Rich skip fusion
Global Attention, high
TransUNet 0.87 0.80
compute
Swin- Local + global fusion,
0.89 0.83
UNet++ efficient
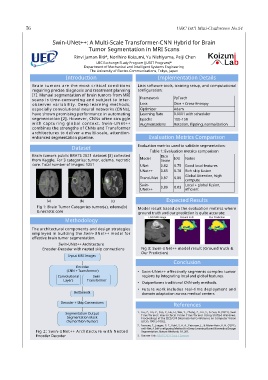
(a) (b) (c) Expected Results
Fig 1: Brain Tumor Categories tumor(a), edema(b)
Model result based on the evaluation metrics where
& necrotic core
ground truth and our prediction is quite accurate:
Methodology
The architectural components and design strategies
employed in building the Swin-UNet++ model for
effective brain tumor segmentation.
Swin-UNet++ Architecture
Encoder-Decoder with nested skip connections Fig 3: Swin-UNet++ model result (Ground truth &
Our Prediction)
Input MRI Images
Conclusion
Encoder
(CNN + Transformer)
• Swin-UNet++ effectively segments complex tumor
Convolutional Swin regions by integrating local and global features.
Layers Transformer
• Outperforms traditional CNN-only methods.
• Future work includes real-time deployment and
Bottleneck
domain adaptation across medical centers.
Decoder + Skip Connections
References
1. Liu, Z., Lin, Y., Cao, Y., Hu, H., Wei, Y., Zhang, Z., Lin, S., & Guo, B. (2021). Swin
Segmentation Output
Transformer: Hierarchical Vision Transformer Using Shifted Windows.
Segmentation Mask Proceedings of the IEEE/CVF International Conference on Computer Vision
(Tumor/Non-Tumor) (ICCV), 10012–10022.
2. Isensee, F., Jaeger, P. F., Kohl, S. A. A., Petersen, J., & Maier-Hein, K. H. (2021).
nnU-Net: A Self-configuring Method for Deep Learning-Based Biomedical Image
Fig 2: Swin-UNet++ Architecture with Nested Segmentation. Nature Methods, 18, 201.
Encoder Decoder 3. Dataset Link: BRaTS 2021 Task 1 Dataset